DNA Walk
Table of Contents
DNA Walk
What is a DNA Walk?
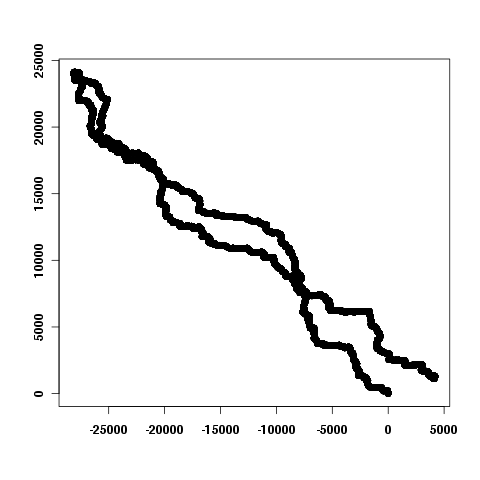
- Read "Genomic landscapes" by Jean R. Lobry for background
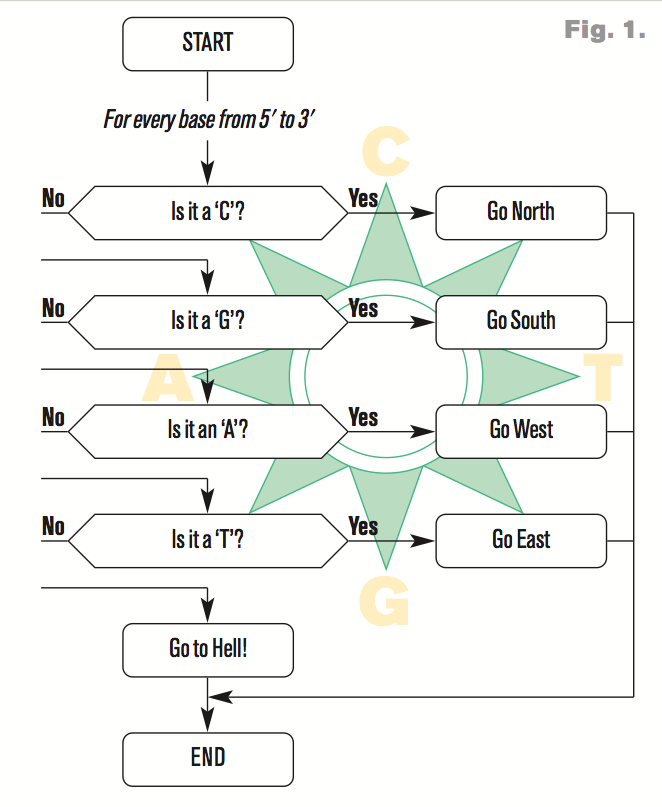
- For every nucleotide, adjust an X or a Y coordinate based on a "compass"
Compass
Project
Overview
- "walking" Borrelia burgdorferi's genome
Big picture
- Get the sequence file into a format Perl can use
- "Walk" the sequence to determine the XY coordinates
- Create a CSV file with the coordinates
- Plot in R or Excel
Input, Output, Process
There are a few different parts to this project.
- Preprocessing (getting the sequence file)
- input
- fasta filename
- output
- array of nucleotides
- Walking the genome
- input
- array of nucleotides
- output
- array of X coordinates, array of Y coordinates
- Creating a CSV file with the coordinates
- input
- array of X coordinates, array of Y coordinates
- output
- CSV file where each row =
X, Y
- Plotting in R or Excel (for completeness)
- input
- DNA walk CSV
- output
- PNG or PDF of the walk
Steps
- Get the sequence file into a format Perl can use (preprocessing)
- open the sequence file
- read the sequence into an array
- close the file
- remove the header line and newline characters
- create an array of nucleotides, e.g.,
('A', 'C', 'G', ...)
- "Walk" the sequence to determine the XY coordinates
- create
@xand@yarrays to hold your coordinates - initialize
$x[0]and$y[0]to 0 (the origin) - for every nucleotide, assign a coordinate based on the compass
- create
- Create a CSV file with the coordinates
- print the coordinates to a CSV
- Plot in R or Excel